
SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets, BMC Research Notes

Standardized phylogenetic and molecular evolutionary analysis applied to species across the microbial tree of life

VDAP-GUI: a user-friendly pipeline for variant discovery and annotation of raw next-generation sequencing data
Opportunistic pathogens and large microbial diversity detected in source-to-distribution drinking water of three remote communities in Northern Australia

Agronomy, Free Full-Text

Metabolism of ʟ -arabinose converges with virulence regulation to promote enteric pathogen fitness
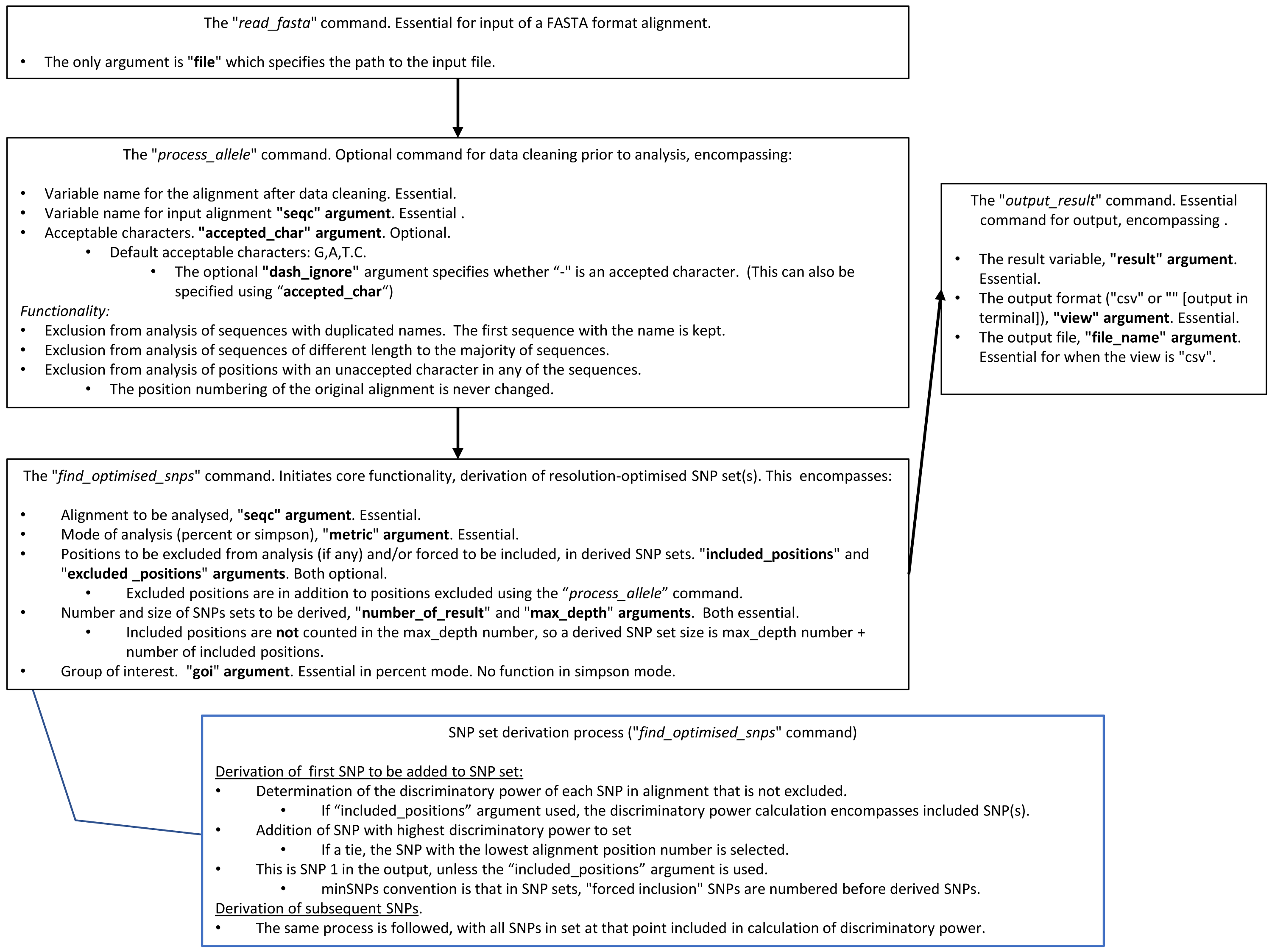
minSNPs: an R package for the derivation of resolution-optimised SNP sets from microbial genomic data [PeerJ]

PDF) Comparative genomic analysis identifies X-factor (haemin)-independent Haemophilus haemolyticus: a formal re-classification of 'Haemophilus intermedius
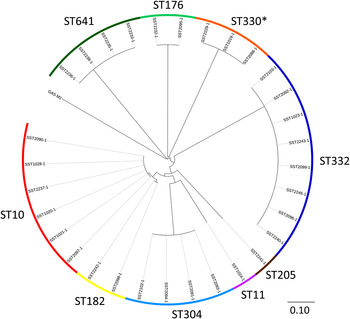
Whole genome sequencing reveals extensive community-level transmission of group A Streptococcus in remote communities, Epidemiology & Infection

Dynamics of genetic variation in Transcription Factors and its implications for the evolution of regulatory networks in Bacteria
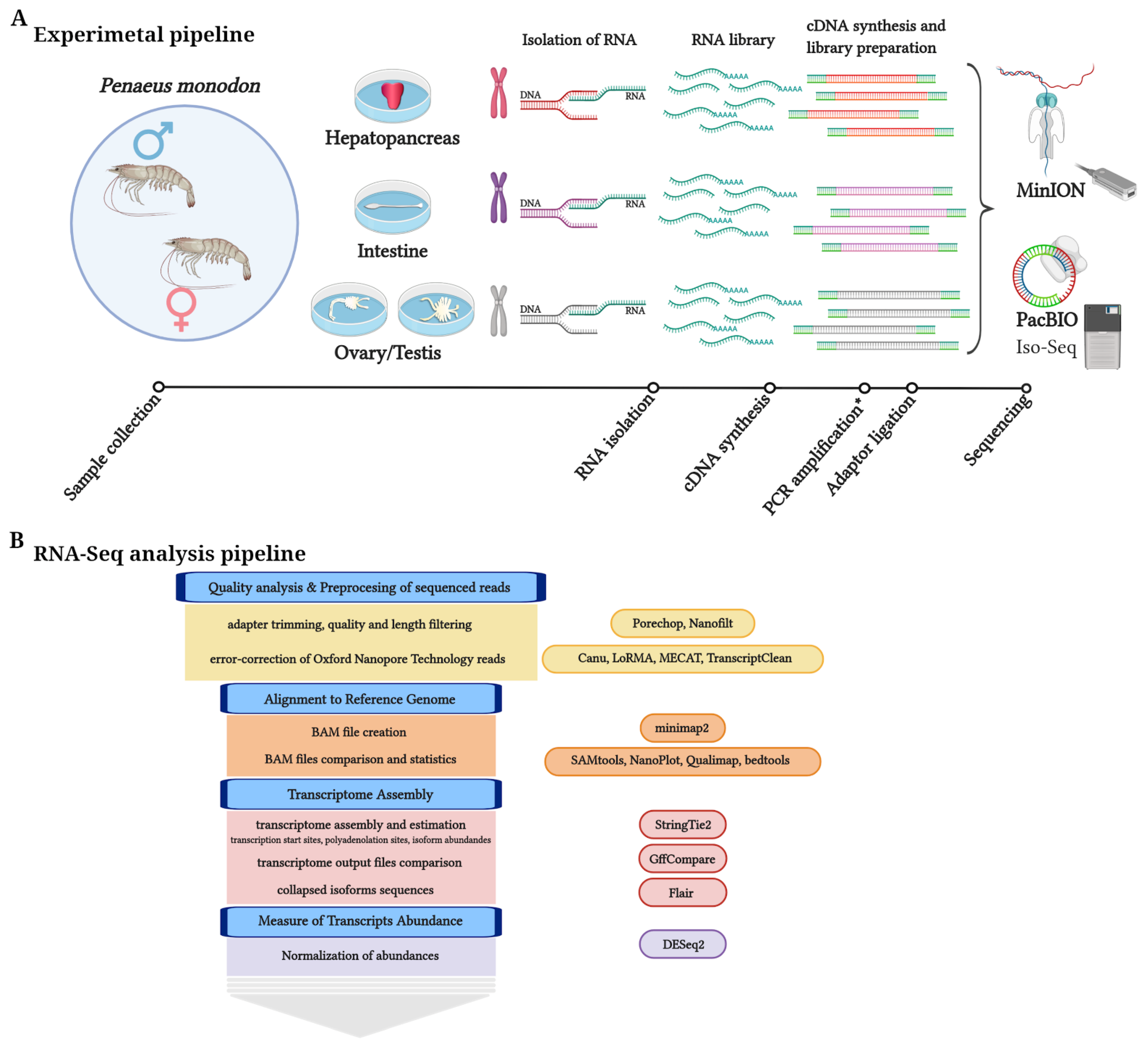
Life, Free Full-Text

Dynamics of genetic variation in Transcription Factors and its implications for the evolution of regulatory networks in Bacteria
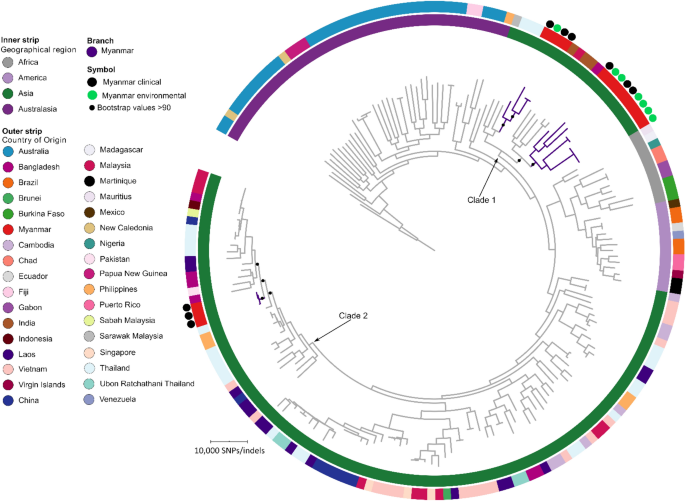
Myanmar Burkholderia pseudomallei strains are genetically diverse and originate from Asia with phylogenetic evidence of reintroductions from neighbouring countries
SPANDx: A genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets - University of the Sunshine Coast, Queensland
Phasing analysis of lung cancer genomes using a long read sequencer

Comparative Genomics and Antimicrobial Resistance Profiling of Elizabethkingia Isolates Reveal Nosocomial Transmission and In Vitro Susceptibility to Fluoroquinolones, Tetracyclines, and Trimethoprim-Sulfamethoxazole








