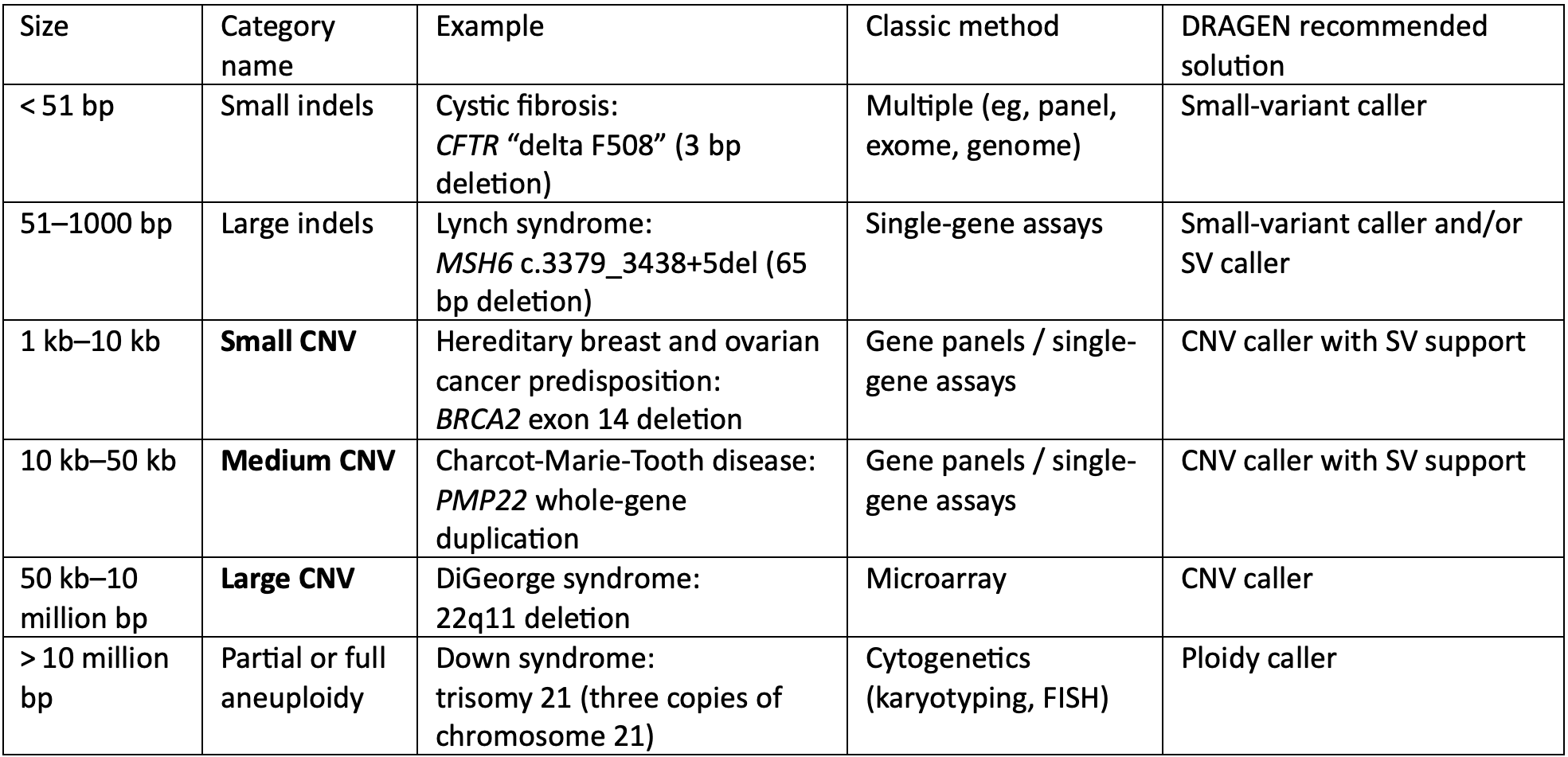
Technical spotlight: Detecting small- and medium-length copy number variants by whole-genome sequencing
Historically, detecting different sizes of genetic variants has required using multiple different tests. By combining Illumina WGS with secondary analysis algorithms built into the DRAGEN Bio-IT Platform, researchers can achieve high-sensitivity detection of all these different variant types using a mixture of methods described here.
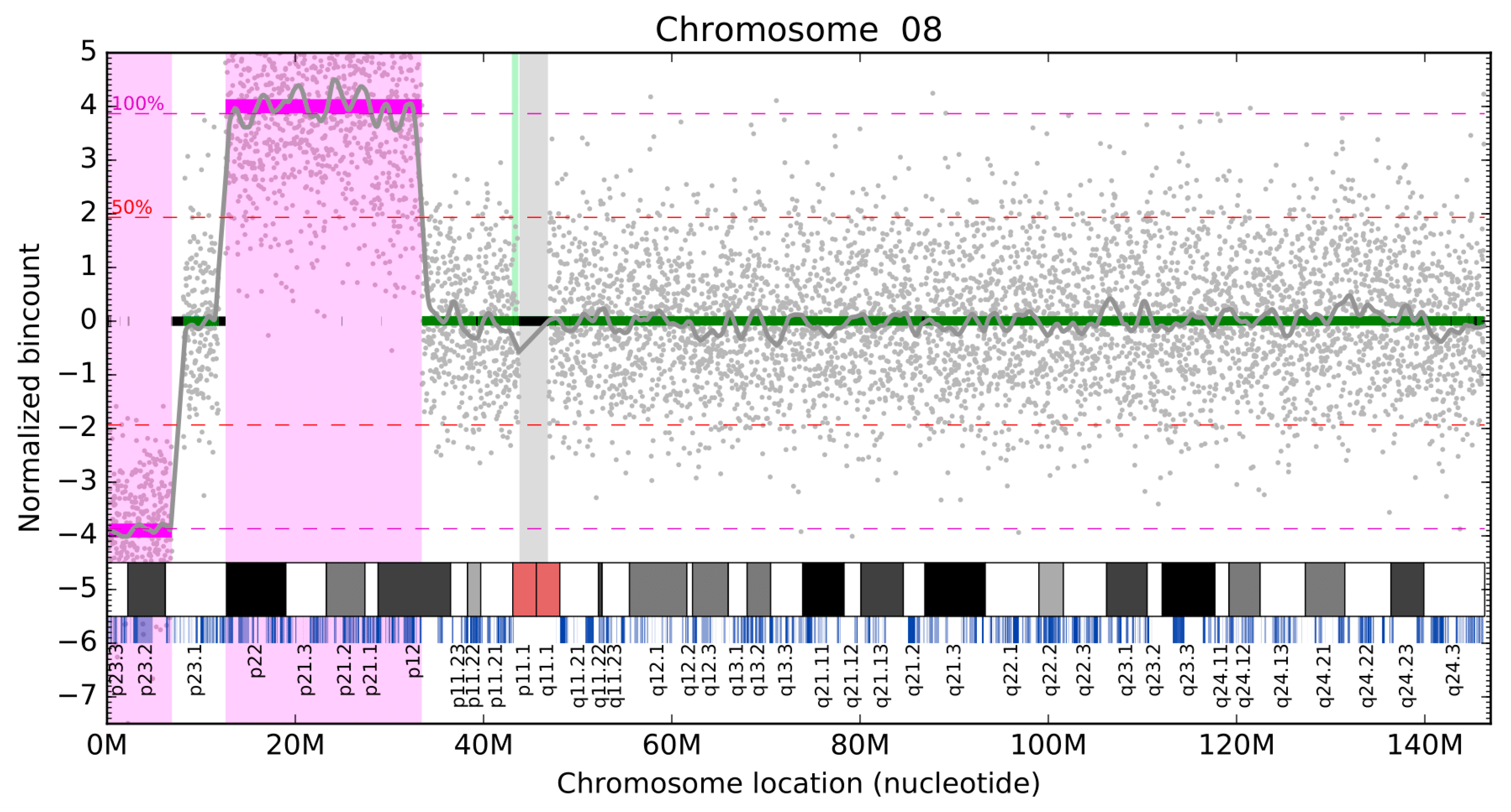
Diagnostics, Free Full-Text

Illumina: The Measurement Monopoly - by Elliot Hershberg

James Han on LinkedIn: Technical spotlight: Detecting small- and

Computational tools for copy number variation (CNV) detection using next-generation sequencing data: features and perspectives, BMC Bioinformatics
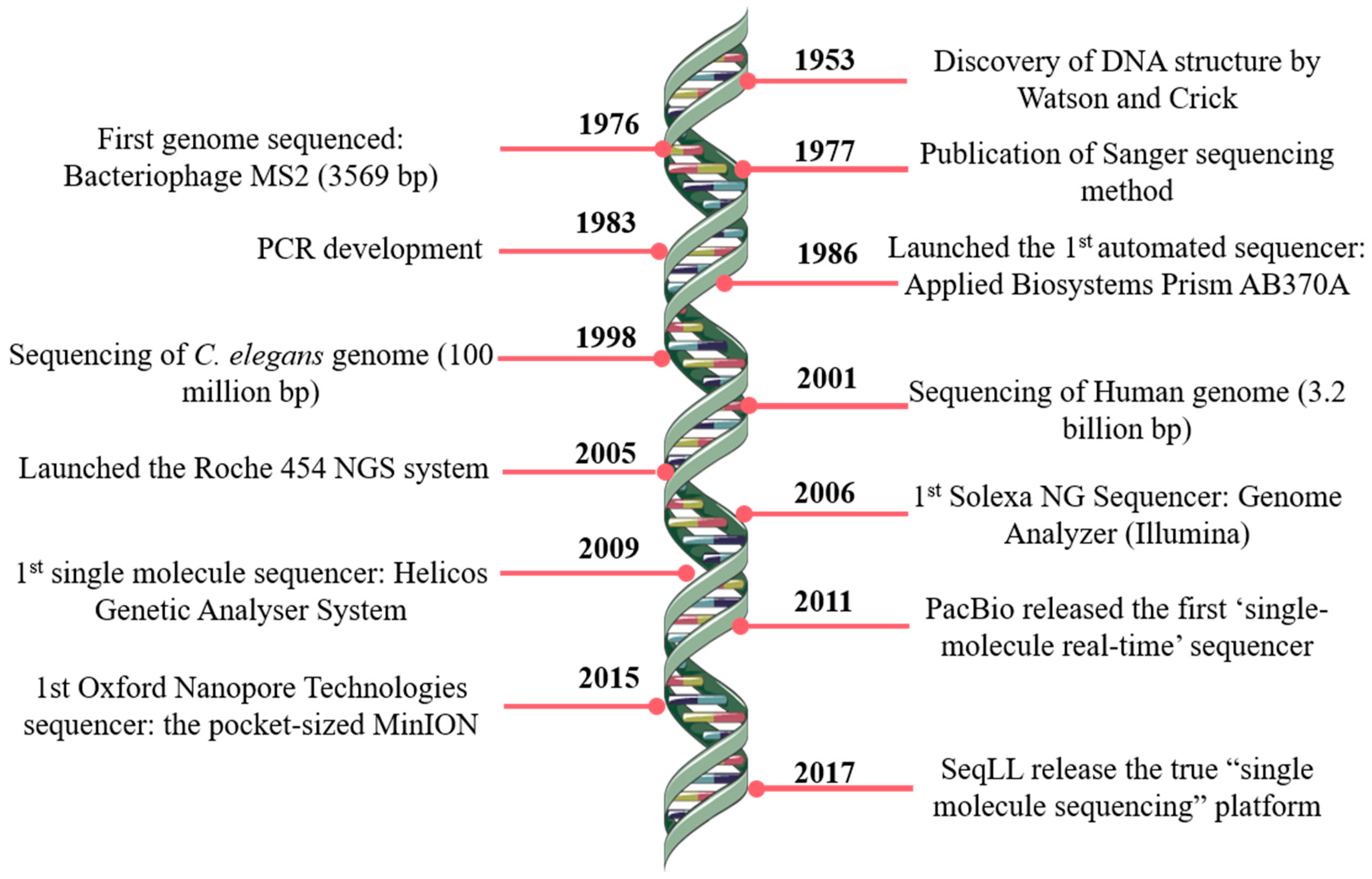
JCM, Free Full-Text

Rami Mehio on LinkedIn: #dragen #fullyfeatured #maximizewgs

Rami Mehio on LinkedIn: #dragen #ukbiobank #wgs #aws #wholegenome
Single-cell copy number variant detection reveals the dynamics and diversity of adaptation

FlowCNV-seq: an almost novel metthod for single-cell copy number analysis - Enseqlopedia

Rami Mehio on LinkedIn: Efficient cloud data analysis for COPD
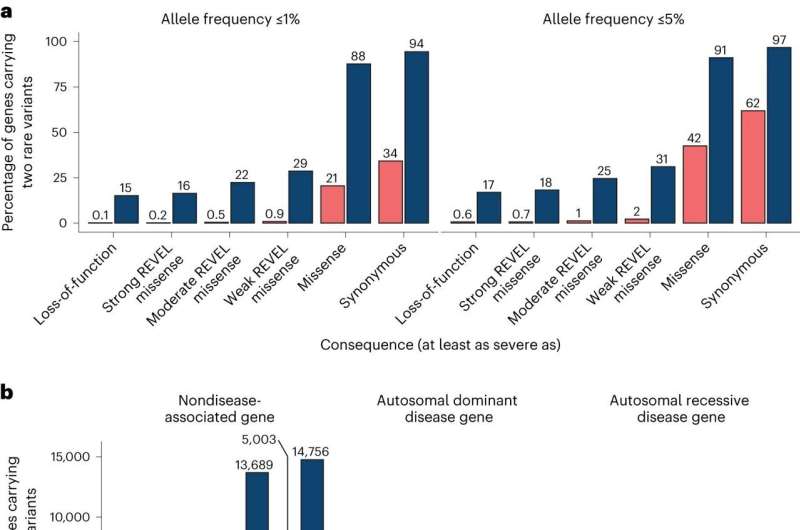
Researchers develop approach to study rare gene variant pairs that contribute to disease

Maria Martínez-Fresno Moreno on LinkedIn: State of California
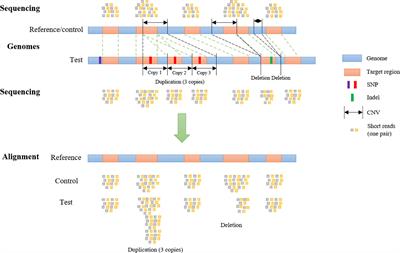
Frontiers SECNVs: A Simulator of Copy Number Variants and Whole-Exome Sequences From Reference Genomes
COVID-19









