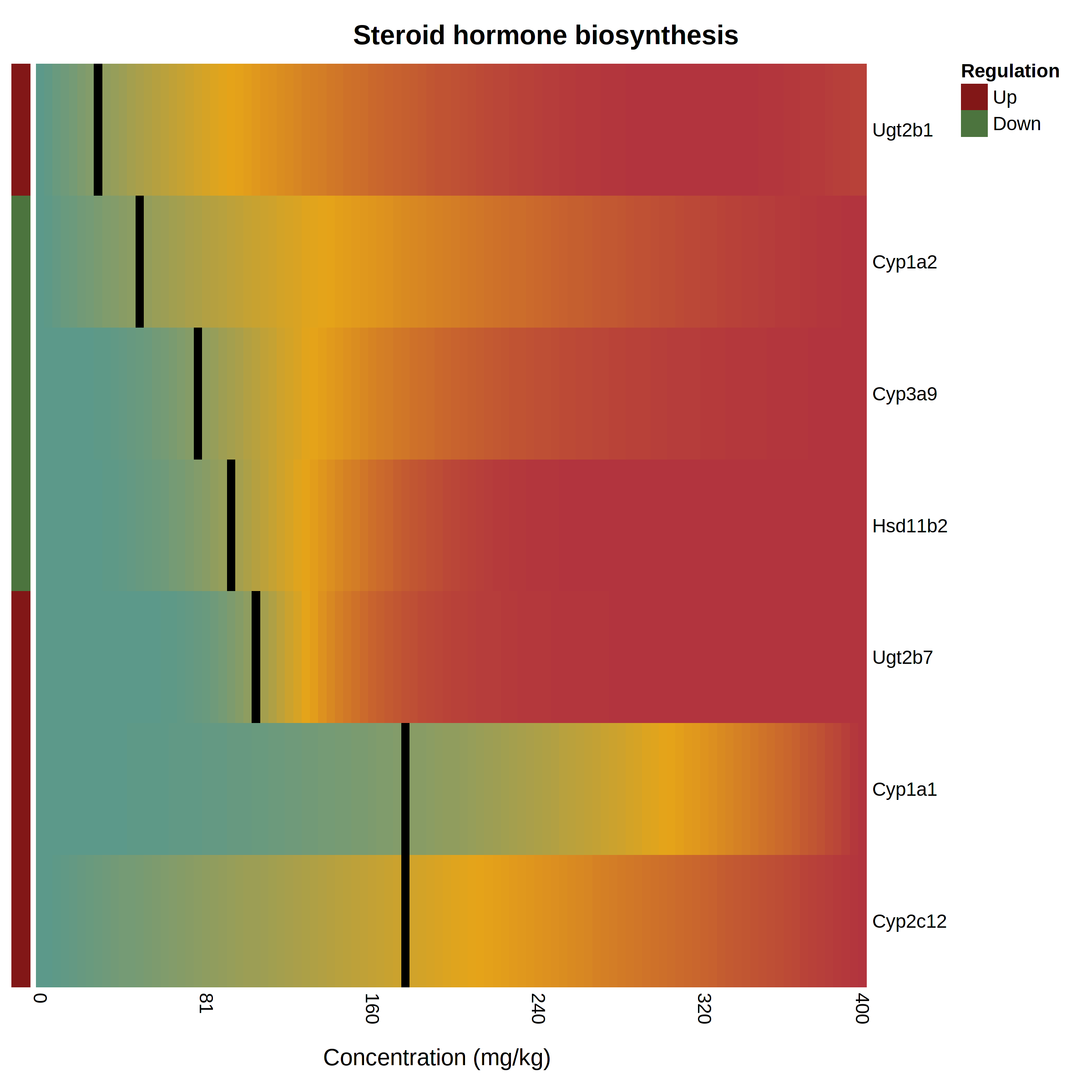
What do the colours on the pathway heatmaps represent? - FastBMD - OmicsForum
The pathway heatmap is an appealing visualization to clearly shows how the expression of each pathway gene compares to the others. It is generated when you click a pathway or gene set name in the “Gene Set Enrichment” panel at the result page. An example output is shown below The pathway heatmap values are calculated through a series of steps: The fitted model for each gene is evaluated across the range of doses in the uploaded data. The resulting modeled expression values are normalized

Heatmap of differentially activated pathways shown in Figure 1A.
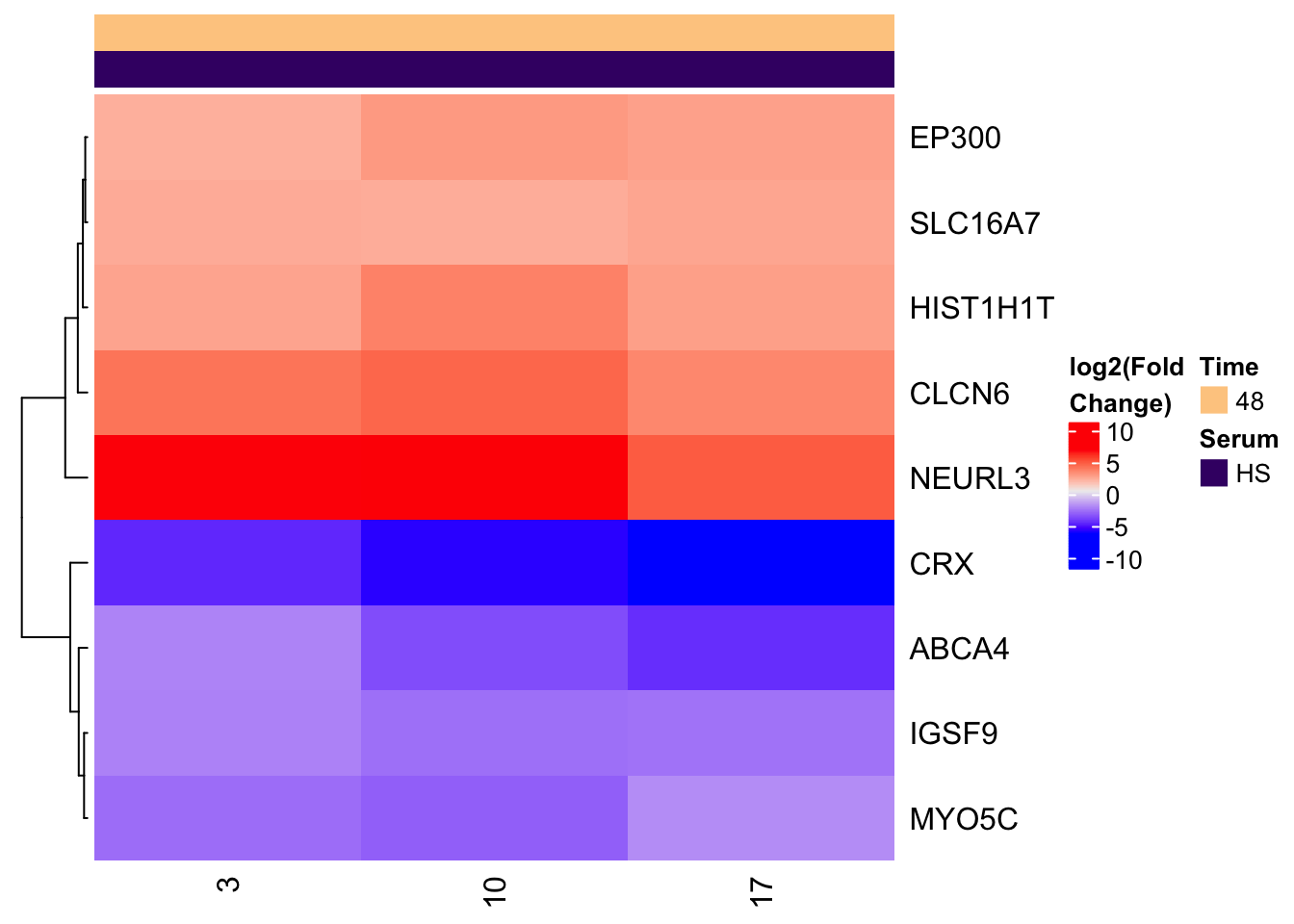
Let's Plot 6: Simple guide to heatmaps with ComplexHeatmaps
d2mvzyuse3lwjc.cloudfront.net/doc/en/Tutorial/imag

Pathway-level heatmap depicts metabolic pathways whose expression was
KEGG Decoder heat map based on Kofamscan results. The heat map
d2mvzyuse3lwjc.cloudfront.net/doc/en/Tutorial/imag

Scientific colour maps - s-Ink · accessible science graphics
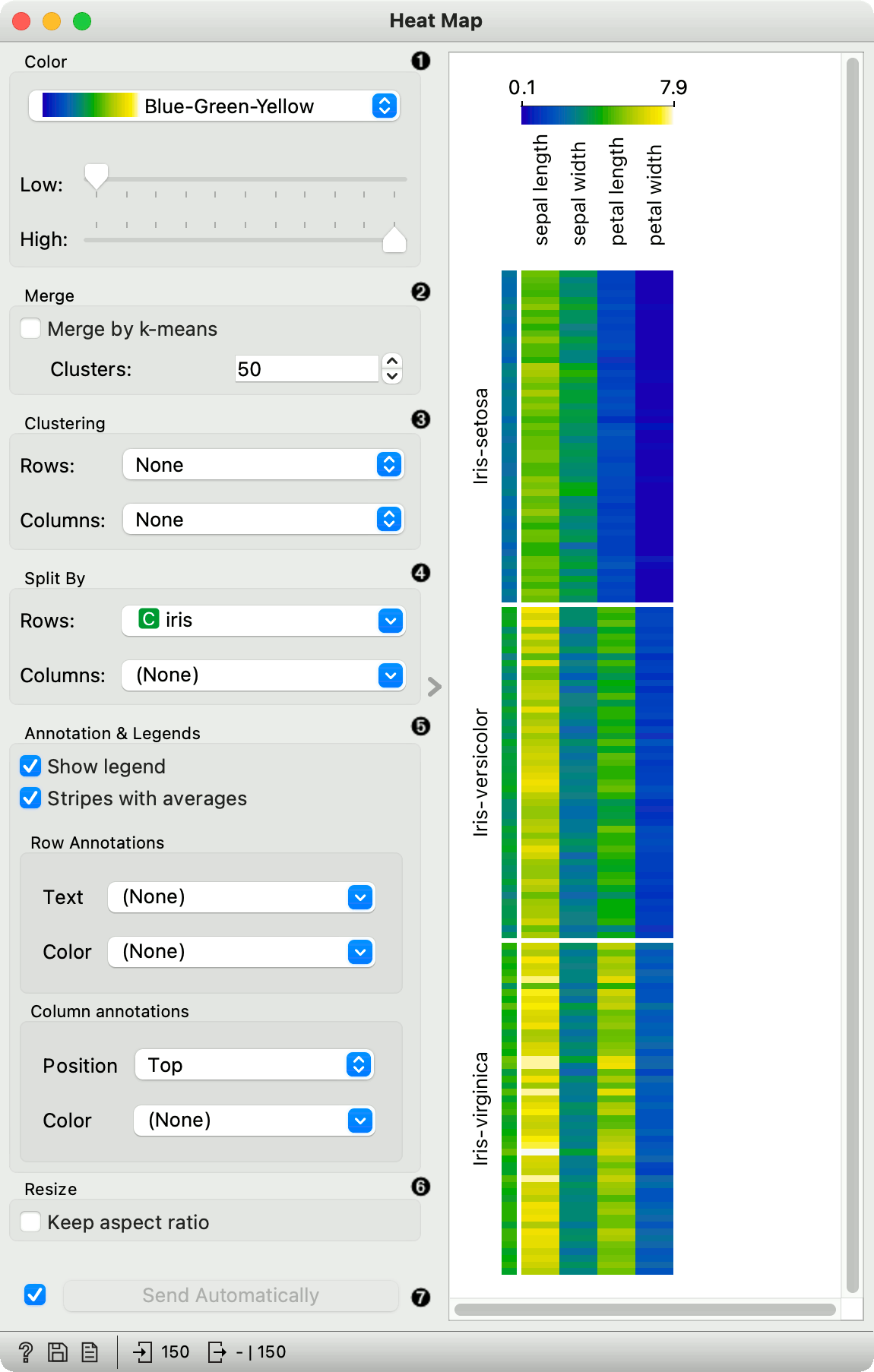
Heat Map — Orange Visual Programming 3 documentation

RAMMB: GOES-R Proving Ground Real-Time Product Demos
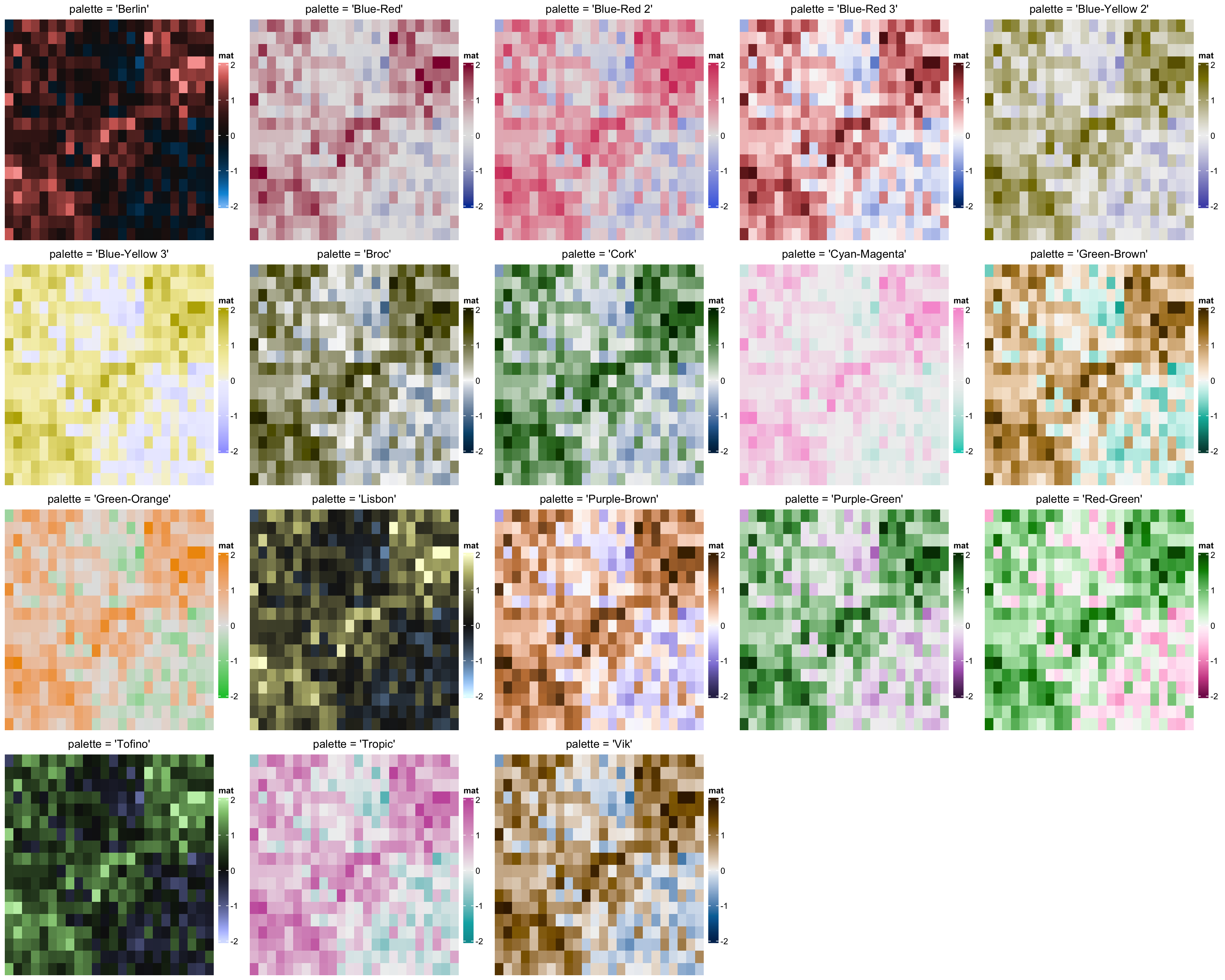
Support HCL colormaps in ComplexHeatmap - Zuguang Gu
Make heatmaps in R with pheatmap - Kamil Slowikowski

Help MetaboMAPS
Knowledge: Comparison analysis heat map for canonical pathways

Heatmap based on the main KEGG pathways predicted at the different









